
^ König, Julian Zarnack, Kathi Luscombe, Nicholas M.This helps to identify PCR over-amplification effects in the high-throughput sequencing step and therefore improves the quantification of binding events. The primer used for reverse transcription can contain a random sequence, which can be used to barcode cDNAs.

The small amount of resulting cDNAs is then PCR amplified and sequenced using a next-generation sequencing platform. Analogous resolution can be obtained with standard HITS-CLIP methods, using the observation that RT that does read through the cross link site has a high error rate, which can be used to deduce the position of the crosslink ("Crosslink induced mutation site" (CIMS) analysis ). The RNA is then reverse transcribed, causing cDNAs to often truncate at the crosslink site, which is the key insight and unique feature in the development of iCLIP, as it allows identification of the site of RNA-protein interaction at high resolution. The radiolabelled protein-RNA complexes are then excised from nitrocellulose, and treated with proteinase to release the RNA, leaving one or two amino acids at the crosslink site of the RNA. As with all CLIP methods, iCLIP allows for a very stringent purification of the linked protein-RNA complexes, using immunoprecipitation followed by SDS-PAGE and transfer to nitrocellulose.
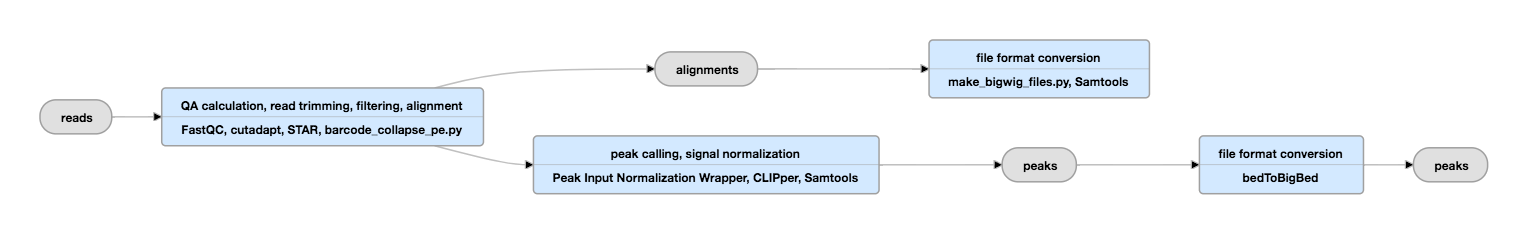
This cross-linking step has generally less background than RNA immunoprecipitation protocols. The method uses UV light to covalently bind proteins and RNA molecules.

ICLIP (individual-nucleotide resolution Cross-Linking and ImmunoPrecipitation) is a method used for identifying protein-RNA interactions. Not to be confused with Intramembrane protease, sometimes abbreviated I-CLiP.


 0 kommentar(er)
0 kommentar(er)
